Published on
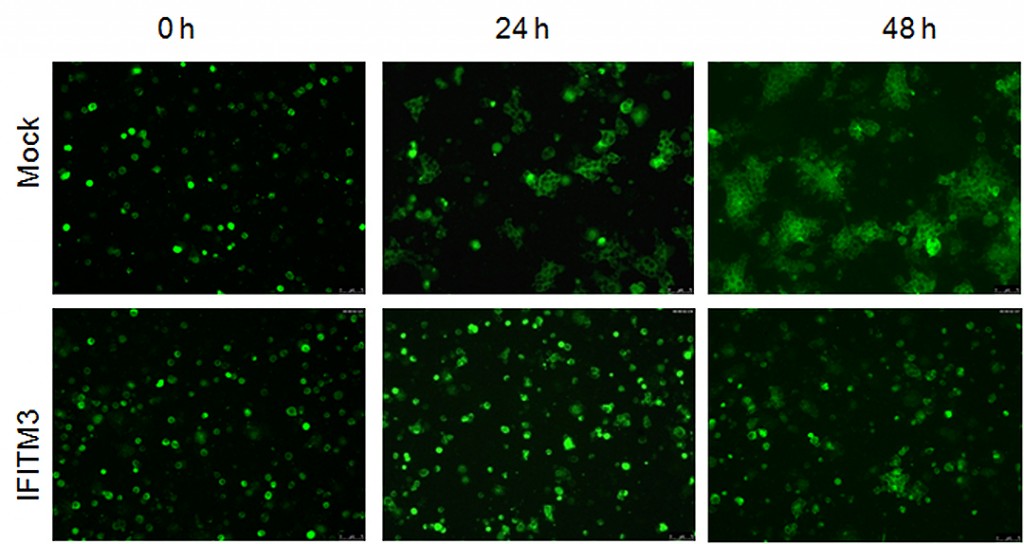
Cells that expressed IFITM proteins (bottom row), showed much less spread of HIV-1 compared with cells lacking the protein. | courtesy Jordan Wilkins, Liu Lab
By Jennifer Lu | MU Bond Life Sciences Center
For Shan-Lu Liu, thinking outside the box meant putting an antiviral protein inside HIV-infected cells, rather than into healthy ones.
Liu and his team of researchers studied how interferon-induced transmembrane (IFITM) proteins limit the infection of HIV-1, the primary strain of virus responsible for AIDS. The journal Cell Reports published their results on September 17.
IFITM proteins are biomolecules with broad antiviral properties. Although multiple versions of IFITM have been found in humans, three are known to have antiviral properties: IFITM1, IFITM2 and IFITM3.
In a 2013 paper published in PLoS Pathogens, the Liu laboratory demonstrated that these three IFITM proteins have the ability to thwart a variety of viral infections.

Shan-Lu Liu, Bond Life Sciences scientist and associate professor in the MU School of Medicine department of molecular microbiology and immunology. | Photo by Justin Kelley, University of Missouri Health Care
“They can inhibit influenza virus, Ebolavirus, HIV and SARS coronavirus,” said Liu, an associate professor in the Department of Molecular Microbiology and Immunology at the Bond Life Sciences Center.
Liu wanted to know why IFITM’s inhibition of HIV was uncharacteristically weaker than its inhibition of other viruses.
To study this conundrum, many researchers designed their experiments by expressing IFITM proteins in target or healthy cells. Then they infected these IFITM-bolstered cells with HIV, but saw minimal protection against viral infection.
In a twist, the Liu group put IFITM proteins in HIV-1 producer or infected cells instead of in healthy T-lymphocyte cells, a special kind of immune cell used specifically to study the viral infection by HIV.
They found that IFITM proteins, especially IFITM2 and IFITM3, interacted with the viral envelope protein (Env) that makes up the outer shells of virus particles.
For normal HIV infections to occur, Liu said, envelope proteins need to be cleaved into two parts.
Once processed, the resulting two portions, Env gp120 and gp41, can be incorporated into viral particles. The two processed envelope proteins protrude from the outer surface of the virus like mushroom-shaped pegs that help the virus latch on and fuse to target cells.
But when IFITM binds to envelope proteins they interfere with the viral envelope functions.
“It’s just unexpected,” Liu said, about this finding. In other viruses his group has studied, IFITM inhibited virus’ ability to fuse its outer shell with the membrane of a cell by making the cell membrane rigid during the infection process. He said he assumed IFITM would block HIV the same way.
Instead, they found evidence suggesting that IFITM blocks infection through direct contact with HIV’s envelope proteins.
“It is the first study that shows this kind of interaction,” Liu said. “That’s why this study is so surprising. We did not think about this.”
Liu does not yet know the mechanism behind IFITM and envelope protein interactions, but he said the outcome remains clear. “IFITM proteins inhibit this Env cleavage process and this makes HIV less infectious and less transmissible.”
To visualize IFITM’s inhibitory effects in action, Liu’s group tagged HIV-1 inside infected cells with a green fluorescent dye. Then they colored healthy target cells with a red fluorescent dye. When they mixed the two populations of cells together, they saw two days later a very tiny amount of cells exhibiting green signals within the red cells —a sign of spread of HIV cell-to-cell infection.
By comparison, cells in the control group—healthy red-tagged cells mixed with green HIV-infected cells that do not contain IFITM—showed a higher number of red cells lighting up green inside.
This suggested that having IFITM and HIV-1 inside the virus-producing cells somehow limited the virus’ infectivity and cell-to-cell spread at the same time.
His group also showed through a technique called co-immunoprecipitation that IFITM proteins bound specifically with envelope protein rather than with other proteins, such as Gag. Liu attributed this work to his two talented hardworking graduate students, Jingyou Yu and Minghua Li.
Unfortunately, the benefits of IFITM are short-lived. When the Liu laboratory let HIV-infected cells replicate again and again, they saw that HIV could evolve enough to circumvent the inhibitory effects of IFITM after 30 passages.
Liu said that his research on IFITM was still in its early stages, but that the next step would be to look at the IFITM’s function in HIV patients in order to move the basic research of IFITM from bench to bedside.
“Once we know better how this protein works, we can develop some inhibitors to block HIV, block Ebola, block other viruses,” Liu said. “So that’s our ultimate goal.”
Liu is a Bond Life Sciences Center investigator and an associate professor of molecular microbiology and immunology in the MU School of Medicine. He studies how viruses infect healthy host cells to cause illness and cell response to viral attacks.
The National Institutes of Health and the Canadian Institutes of Health Research partially supported this research. Additional collaborators include Eric Freed, PhD, senior investigator with the National Cancer Institute (NCI) HIV Dynamics and Replication Program; Chen Liang, PhD, at McGill University; and Benjamin K. Chen, PHD, at the Mount Sinai School of Medicine. Read the full study on the Cell Reports website and browse the supplementary data for this work. See more on this research from Mizzou News.