Published on
Updated on
Efforts to understand the genome of one plant through its many genetic varieties could lead to nutritional improvements in the staple crops billions of people depend on
By Phillip Sitter | Bond LSC
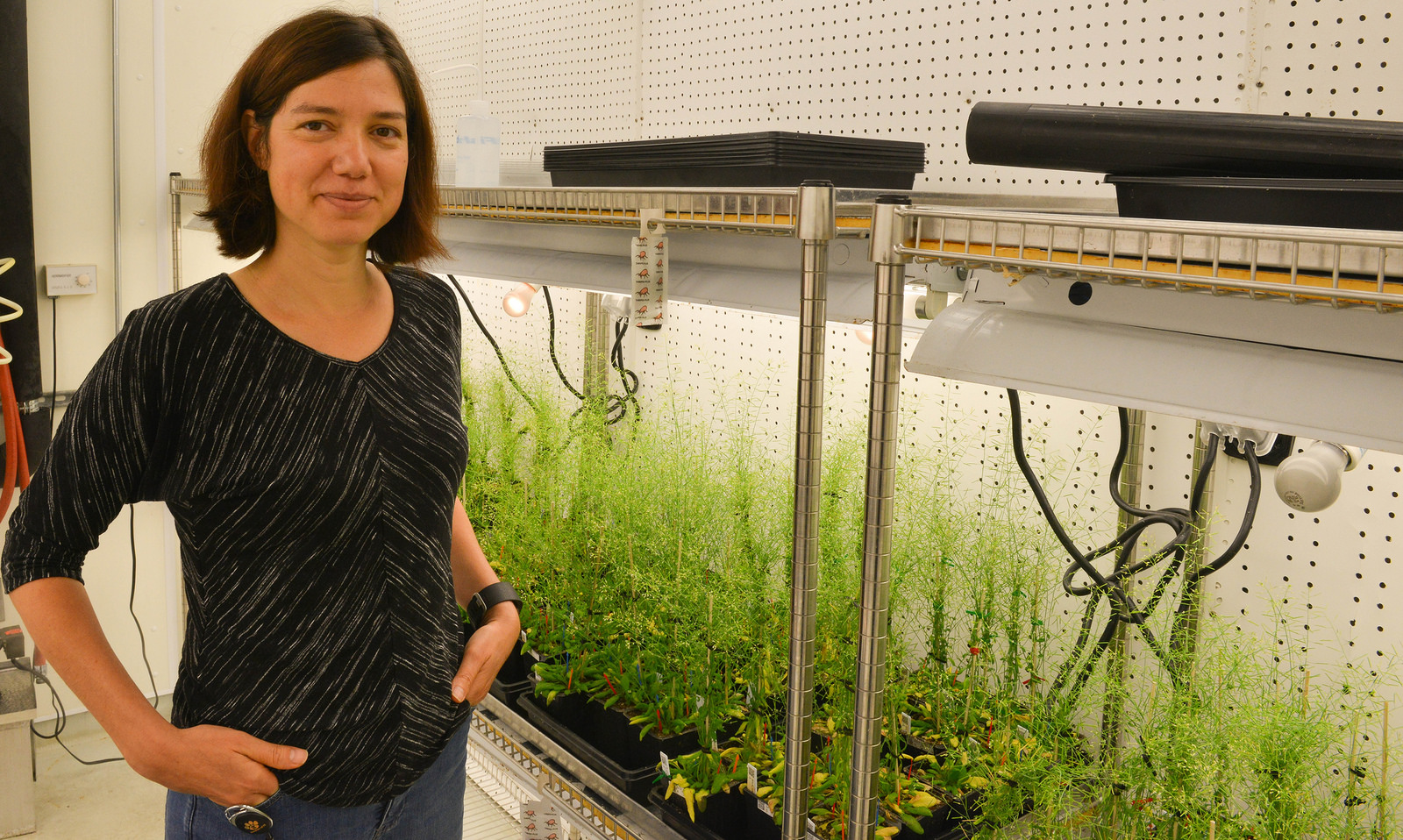
Ruthie Angelovici stands next to some Arabidopsis thaliana samples in the basement of Bond LSC. She is leading projects to study the relationships between genotypic and phenotypic variation in Arabidopsis and how this affects the amino acid content of the plants, and the resistance of their seeds to drought conditions. | Phillip Sitter, Bond LSC
It’s hard to avoid corn, rice or soybeans in your diet, and you’ve probably eaten or drank something today with at least one ingredient from them.
Unfortunately for the billions of people worldwide who depend on these crops as a staple, they aren’t actually all that nutritious. Specifically, they lack sufficient quantities of amino acids.
Twenty amino acids are required to build any protein, and within that about ten are considered essential, Bond Life Sciences researcher Ruthie Angelovici said. “Without amino acids, you can’t live.”
Amino acids might seem minor, but important parts and processes in our bodies from our muscles to enzymes are built from or work through them. That’s why Angelovici wants to enhance their availability in key foodcrops.
In the case of amino acids, “What we’re trying to understand is the basic question of how those accumulate in seeds, and then from that basic concept we’re going to try to improve that in grain,” Angelovici said.
The evolution of poor nutrition
No one really knows why so many of our most important crops that essentially sustain humanity lack sufficient essential amino acids.
Maybe plants don’t synthesize amino acids because the cost in energy for the plant is too high, or because higher levels of amino acids might make them more vulnerable to attacks from hungry insects. Maybe if plants produced higher levels of amino acids, the taste would be too strong for human palates, and so our ancestors long ago selectively bred those traits out of crop populations. Or, maybe in ancient farmers’ pursuits of other traits in their crops, like higher quantities of starch, humanity accidentally boosted one nutritional trait at another’s expense. There are a lot of unknowns when it comes to these theories, Angelovici said.
What is clear — and something Angelovici said she cannot stress enough — is how powerful a genetic tool she and her fellow researchers at Bond LSC have in the form of a collection of a vast amount of genetic variation of Arabidopsis thaliana.
“Arabidopsis thaliana is a model [plant] system that a lot of plant scientists use, although it is not a crop, or anything like that, but it’s a great model plant to start with, and then everything we learn from it, we can try and figure out if it’s the same in maize, rice, soybean, and translate it,” Angelovici explained.
Part of the mustard family, Arabidopsis grows quickly so researchers can study four or five generations in one year. As an added bonus, this huge genetic variety but can be grown in just one room instead of large fields. For Angelovici, that room is in Bond LSC’s basement and the basement of greenhouses nearby.
“We are growing right now 1,200 ecotypes of this Arabidopsis thaliana. So, what is an ecotype? It’s basically from the same species, but they have a slightly different genotypes. So, we’re looking at a vast genetic variation that represents genetic variation of this species across the world. Each ecotype comes from a different place,” she said.
For those of you wondering, a genotype is the specific sequence of information in an organism’s genetic code — its genetic identity. A phenotype is an observable physical trait controlled by the genetic sequence. For phenotype, think in terms of color, size, shape — just like in different breeds of dogs and cats, for example.
Even the smallest differences in genetics can produce the range of traits we observe, like the size difference between a Chihuahua and a St. Bernard — even though all the breeds are the same species. The same thing applies to plant species, too.
Angelovici said researchers can use all the genetic variation in their extensive Arapidopsis collection understand questions of how observable traits relate to genes, and vice versa.
Once that connection is established, “we basically have an address on the genome, and then we can go after the gene itself, understanding the function of the gene, and how that is affecting our variation of the phenotype, basically to help us understand the mechanism,” Angelovici explained.
“And if you understand the mechanism, we might be able to improve it, change it, either through genetic engineering or breeding. Basically, mining what Mother Nature has already done throughout many generations, and trying to figure out if we can utilize that in crops,” she added.
“We can measure the level of amino acid, but does the plant really care about the absolute level of amino acid, or relative level, and how they correlate with one another? It appears that these relationships are really important.”
All this algorithmic analysis can eventually improve results.
“When we get a candidate gene that we think affects one of the traits that we are interested in, we either knock it out or over-express it, and go back to the phenotype and figure out if it changes, and how,” Angelovici said.
“Along the way, we also try to understand if the phenotype is correlating with something that is larger, for example the plant’s growth, its development or the development of seeds.”
A plant under stress
An understanding of seed development might be especially important in understanding how drought affects the nutritional quality of future generations of water-stressed plants.
“Surprisingly, those are processes that are not well-understood — how the seed itself is adapting to water stress. A lot of people are working on water stress and drought at the plant level, in the yield [of a crop], but we’re trying to really understand what is happening on the level of the seeds, on the bio-chemical level, and then how that affects the next generation,” Angelovici explained.
If she and her fellow researchers find a super-resilient seed, they could learn to transfer its resiliency to drought to future generations of seeds.
Something they’ve seen already is that if you really water-stress a plant, while it may produce less seeds, seeds that it does produce are bigger.
“Right now the question is, are they bigger because they are trying to adapt for their harsher environment, or are they just trying to survive?” she said. Is the parent developing its offspring in a certain way to ensure the best possibility of success of that offspring, or just so it can survive to reproduce another day?
“We can only provide the data,” Angelovici said of her work in trying to answer questions like these, in order to improve the quality of human life by understanding and improving the quality of our food.
“This is the mechanism, and that is a tool we can provide,” Angelovici said of what the research can offer to people like farmers and other plant breeders. “Knowledge is power. What we do with this power is up to a lot of people.”
Ruthie Angelovici is an assistant professor in the Division of Biological Sciences, and is a researcher at Bond Life Sciences Center. She received her degrees in plant science from institutions in Israel — her B.S. and M.S. from Tel Aviv University, and her Ph.D. from the Weizmann Institute of Science in Rehovot. She was a postdoctoral fellow at the Weizmann Institute and at Michigan State University, and has been at MU since fall of 2015.

This file photo shows at least one other ecotype of Arabidopsis thaliana in a greenhouse in Bond LSC. Even small variations in the species’s genome can create the large number of observable varieties, sometimes with distinct sizes and shapes, other times with genetic differences that can only be observed on the microscopic level. | Roger Meissen, Bond LSC