Published on
Updated on
Research illuminates how one of the most prevalent zoonotic diseases infects cells

By Sarah Gassel | Bond LSC
When bacteria invade a host, they employ unique strategies to weaken the host’s cells for optimal infection. For the bacterial pathogen Brucella, this means manipulating internal cell machinery to subvert host function and favor infection.
New research at the University of Missouri and Texas A&M University reveals a specific mechanism not previously observed that this major public health concern uses to achieve this takeover. Their study published in the journal Cell Host & Microbe March 25, 2024.
“Brucella is a very smart pathogen, and a lot of how it manipulates a host’s function is largely unknown,” said Qingming Qin, a study author in the lab of Paul de Figueiredo, a principal investigator at MU’s Bond Life Sciences Center.
Brucella causes brucellosis, one of the most common zoonotic diseases worldwide. More prevalent in resource-limited countries, it primarily infects cattle, pigs, goats, sheep and dogs, but humans can contract the disease through eating or drinking unpasteurized milk or cheese from the animals. Brucellosis has symptoms similar to malaria and infects an estimated 2.1 million people globally each year, according to the Centers for Disease Control. The frequency of illness relates to the pathogen’s complex mechanisms for infection. In addition to the public health concerns raised by brucellosis, its economic impacts are also a major driver of poverty in developing countries.
Generally, pathogens are able to evade recognition by the immune system in a host cell by disrupting a cell’s processes. They do this by targeting specific cell parts with necessary roles, such as proteins and sugars. Sugar molecules, called glycans, are building blocks of cell components. In addition to providing the cell with energy, glycans allow these components to function properly.
Because of their widespread role throughout the cell, sugars are an ideal target for manipulation by bacteria. De Figueiredo joined with researchers at Texas A&M, Texas Tech, CIRAD (French Agricultural Research Centre for International Development) and other colleagues to reveal a new mechanism bacteria use to do this.
“Our results demonstrate the potential of systems biology to enhance our understanding of bacterial ultimate adaptation to their hosts and to imagine new therapeutic tools,” said Damien Meyer, a study author and principal investigator at CIRAD UMR ASTRE.
Many bacterial pathogens have secretion systems, which operate like syringes that can inject material into a desired host cell. In the case of Brucella, it injects proteins into the cell to manipulate the cell’s machinery.
Known as an effector protein, it influences the cell’s systems to weaken and allow for successful bacterial infection and reproduction. Different effector proteins affect different parts of cell systems, which gives researchers insight into how to hinder a bacterial invasion.
“Studying the mechanisms underlying Brucella-host interaction can provide us a lot of new insight on how Brucella can infect the host cells and find a strategy of how to prevent infection and — even after infection — how we can stop it,” said Qin. “That’s our long-term goal.”
Using software to predict effector proteins, researchers made progress in this goal when they discovered Rhg1, an effector protein not previously identified.
To reveal the function of Rhg1, scientists performed tests that showed Rhg1 interacting with proteins associated with the oligosaccharide transferase (OST) complex. The OST complex is cell machinery that helps produce proteins by adding N-glycans — sugar molecules that specifically modify proteins. N-glycans attach to specific sites on the amino acid chain that makes a protein to determine how it folds. Proteins folded differently carry out different tasks, making N-glycans essential for the overall functioning of the cell. The amino acids are not processed with N-glycans pre-attached; rather, this is completed by the OST complex which catalyzes the transfer and attachment of N-glycans to the amino acids. According to the study, the Rhg1 effector protein targets the OST complex, manipulating these specific N-glycan sugars.
This novel discovery gives additional insight into Brucella’s complex strategies.
“It opens a lot of doors to look at different connections,” said Qin.
This finding also allowed researchers to gain a better understanding of the protein malfunctions that occur in the host cell during infection.
Researchers observed that Rhg1’s modification of the OST complex causes an overall reduction of N-glycan sugars. This deficiency within the amino acid chains results in misfolded proteins that cannot be utilized. The accumulation of unfolded or misfolded proteins in the lumen of the endoplasmic reticulum (ER) compartment causes a cell response known as the unfolded protein response (UPR). During this process, defective proteins are chopped into small blocks that are recycled under normal conditions.
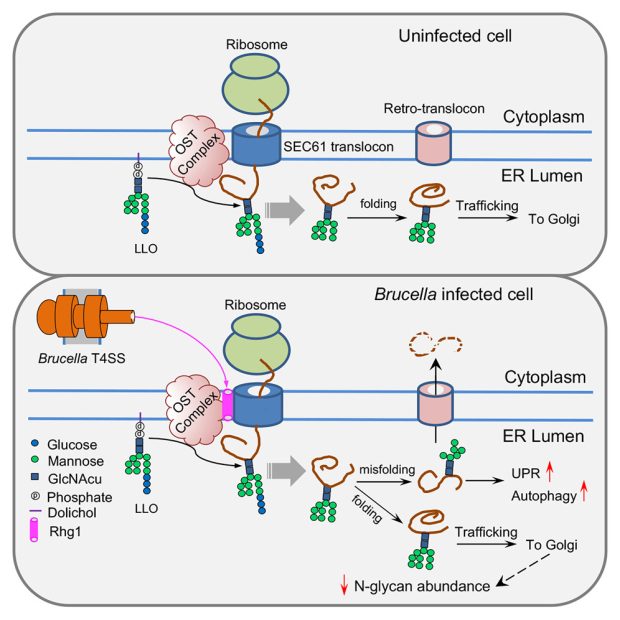
But, Brucella bacteria use this response to their advantage. They use this recycled material for replication, suggesting the bacteria purposefully seek to activate the response. Because of the misfolding, Brucella benefits even more when cells reduce specific proteins that recognize pathogens, which may also help the bacteria avoid activating immune responses from host cells.
Observing Rhg1 revealed to scientists how the pathogen may be smarter than previously thought, and expanding understanding of this infection process could pave the way to prevent human brucellosis by improving current treatments.
“We definitely would like to make good use of this discovery one day in the future,” Qin said.
Cell Microbe and Host journal published “Brucella-driven host N-glycome remodeling controls infection” March 25, 2024. Co-authors include by Ana-Lucia Cabello, Kelsey M. Wells, Wenjing Peng, Hui-Qiang Feng, Junyao Wang, Damien F. Meyer, Christophe Noroy, En-Shuang Zhao, Hao Zhang, Xueqing Li, Haowu Chang, Gabriel Gomez, Yuxin Mao, Kristin L. Patrick, Robert O. Watson, William K. Russell, Aiyung Yu, Jieqiang Zhong, Fengguang Guo, Mingqian Li, Mingyuan Zhou, Xiaoning Qian, Koichi S. Kobayashi, Jianxun Song, Suresh Panthee, Yehia Mechref, Thomas A. Ficht, Qing-Ming Qin, and Paul de Figueiredo.
This work is supported by grants from Texas A&M Clinical Science Translational Research Institute, the Defense Advanced Research Projects Agency, the NIH, the National Science Foundation, the Bill and Melinda Gates Foundation, the University of Missouri NextGen Precision Health Endowment and the National Institute of Child Health and Human Development.