Published on
By Lauren Hines | Bond LSC
With two laptops in front of him and a supercomputer on the edge of campus, graduate student Skyler Kramer runs through code daily in the Dong Xu lab with a purpose far beyond deciphering lines of data. He helps his colleagues defeat cancer.
Working in Bond LSC with senior post doctorate David Porciani from the Donald Burke lab, Kramer and the Dong Xu lab are part of an effort to target cancer tissue and develop more precise delivery of treatment.
In addition, they’re trying to learn more about the biology that drives cancer growth and what makes cancer tissue resistant to treatment after a certain point in time.
“[Kramer] has been a tremendous asset for the project,” Porciani said. “His contribution is so critical at many levels.”
Porciani set out four years ago to find a way to easily differentiate between cancer and healthy tissue. By developing small pieces of RNA and DNA called aptamers that can bind to receptors on cancer cells, he slowly tried to determine the pieces that would only interact with cancer — not bothering the healthy cells — that could deliver a cancer-killing drug. But the process meant Porciani would amass mounds of data.
“There is a lot of statistics in there, which is very important,” Porciani said. “So, that brings in the collaboration with Skyler.”
About a year ago, Porciani was presenting his work on aptamers at a talk in Bond LSC. Xu later inquired if Porciani was open to any collaboration, which led to discussion on what the two labs could do together.
“It was just really really fascinating,” Kramer said. “So, I was really interested in it.”
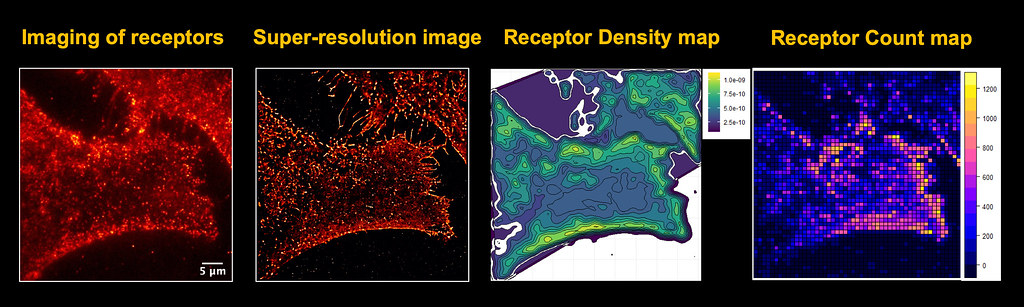
Now, the two are working together to turn microscopy images into something more meaningful.
Through super resolution microscopy images of the receptors on the cancer cells, Porciani took a short video showing the thousands of receptors shaking.
“Most of the receptors like to shake hands with each other, and when that happens it is critical for the growth of the cells,” Porciani said. “So, the handshakes are sort of a signal that says, ‘Okay, let’s move this house to the next stage. Yes, let’s make the cell grow even more.’”
Porciani could analyze only a small part of these shakes with his naked eye but using a bioinformatics approach —using software and statistics to understand biological data — Kramer can take that data from the thousands of receptors and turn it into digestible and meaningful information.
“[I’m] trying to take the massive amounts of data that we’re able to generate and get some sort of useful information out of it,” Kramer said. “So, a lot of times it’s useful not only to get some sort of numeric output but some sort of visualization also.”
This means turning an image of a cancer cell into a receptor density map or a receptor count map. By doing so, researchers can investigate why receptors clump into one area, what that means and why cancer cells become resistant to treatment over time.
“It’s our job to understand why there is this difference,” Porciani said. “This type of information can generate new hypotheses that we need to test.”
With the help of bioinformatics, Porciani and other researchers can find the answers to their questions.
“I think [bioinformatics] is hugely impactful,” Kramer said. “A lot of times, the life sciences will give you the actual experiments and a rough idea of what happened in the experiment, but I think that when you apply the bioinformatics you get more to why specific things are happening.”
The collaboration brings more than just new results, but also can help expand science.
“I think I’m learning a lot from him,” Porciani said. “I think, hopefully, he is learning a lot from me as well.”
To learn more about Porciani and Burke lab’s findings regarding aptamers, the research article can be found here.